Dr Sue Krause
- Research Associate (Molecular Biosciences)
telephone:
01413306811
email:
Sue.Krause@glasgow.ac.uk
pronouns:
She/her/hers
Ri: Mc&Sb, Lab 331 Davidson Building, Glasgow G12 8QQ
Biography
My research career started as an undergraduate student at the University of Wisconsin-Milwaukee, where I received a BS in Biological Sciences. I worked as an undergraduate research assistant in the Department of Chemistry there in the lab of Prof. Stephen Ragsdale. One aspect of the time in the lab was to perfect transformation protocols of E. coli.
I went to the Department of Biology at Johns Hopkins University for my PhD studies. After rotations and being a teacher assistant for two lab classes, I started the research aspect of my degree with Prof. Margarete M.S. Heck. The focus of my PhD thesis was the formation of mitotic chromosomes in Drosophila melanogaster.
My research involved two different approaches:
(1) generating antibodies against proteins associated with isolated mitotic chromosomes
(2) using known Drosophila mutants with abnormal mitotic chromosomes.
During my time with Margarete, we moved to the University of Edinburgh. I fell in love with Scotland (and a Scotsman).
After completing my PhD, I started research with Prof. Joe Gray at the University of Glasgow in the Division of Molecular Genetics. The focus of our research with the better understanding the PKC1-signalling pathway in budding yeast using a mixture of genetic and cell biological approaches. This pathway is involved re-building the yeast cell wall due to various stresses as well as normal building of the cell wall.
After my research with Joe ended, I returned to Drosophila research with Prof. Julian Dow. With Julian, my work has concentrated on Big Data projects: Fly Atlas 2 and Fly Met. Fly Atlas 2 is an active web resource that illustrates the transcriptomics of various Drosophila tissues from male and female adult flies and within the third instar larvae. Fly Met is an active web resource of the metabolomes of the same tissues found in Fly Atlas 2.
Our current research involves a comparison of renal systems of insects using single cell technology.
Research interests
Mitotic chromosome formation requires DNA replication.
My characterization of two known Drosophila mutants that alter the formation of mitotic chromosomes resulted in mutations in two subunits of two different DNA replication complexes.
In the paper, https://doi.org/10.1128/MCB.21.15.5156-5168.2001, we showed that the DNA replication factor c, subunit 4 (Rfc4) is a required protein as mutations in the gene result in larval death. Observing the mitotic chromosomes, we saw abnormal mitotic chromosomes formed. Rfc4 does not localize to mitotic chromosomes itself, thus the impact on the formation of mitotic chromosomes and progression through mitosis is due to interrupting an upstream event. We suggest that the aberrant chromosomes are due to failure to recognize a cell cycle checkpoint that responds to DNA replication inhibition and/or DNA damage.

In the paper, https://doi.org/10.1016/S0960-9822(00)00844-7, we showed that the subunit 2 of the Drosophila Origin of Replication Complex (Orc2) is required for proper mitotic chromosome formation. We showed that mutations in this gene resulted in abnormal timing of DNA replication. We saw areas of euchromatin replicating after heterochromatin DNA in the Orc2 mutants; normally all euchromatin DNA replicates before heterochromatin DNA.

Pkc1 signaling pathway in Saccharomyces cerevisiae
In my first work with Prof. Gray, https://doi.org/10.1016/S0960-9822(02)00760-1, we showed that the Pkc1-signalling pathway responds to disruption of the TOR pathway (Western blot shows Mpk1 activation when yeast cells were treated with rapamycin).

The viability of the yeast mutants in the Mpk1 and Pkc1 genes is severely hampered when treated with rapamycin. We showed that decreased viability was due to an increase in cell lysis. From this study, we inferred that the cell growth that TOR signaling regulates requires the Pkc1-signalling pathway to remodel the cell surface of the yeast cells.

This work led to Joe and I writing a BBSRC grant to identify upstream regulators of Pkc1 in yeast.

With collaborators at the University of Toronto, we used a system they pioneered to identify genes that interact with Pkc1 in a near-genome-wide methodology (synthetic genetic array analysis (SGA)).
We, then, used chemical genetics to confirm those that interacted with Pkc1, by using a known chemical inhibitor of Pkc1. We discovered a novel gene that functioned upstream of Pkc1 – Ydl203c that we named Ack1. This work was published here: https://doi.org/10.1016/S0960-9822(02)00760-1.
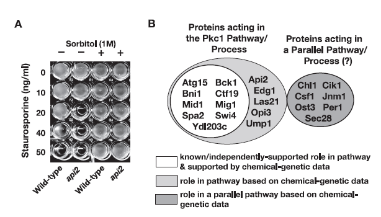
This work led to another grant on which I was the co-Investigator on from the BBSRC.
This grant allowed us to show that Ack1 interacts with the Rho1-GEF, Rom2. We, also, identified another protein that interacted with the other yeast Rho1-GEF, Tus1. Also, while Rom2 and Tus1 act on the same Rho-GTPase, they direct Rho1 to act differently by recruiting it to different places within the cell as was reported in https://doi.org/10.1242/jcs.100685. These graphs illustrate the two Rho-GEFs acting differentially on Rho1. As indicative of two different roles for Rom2 and Tus1, they localize differently within the yeast cell.
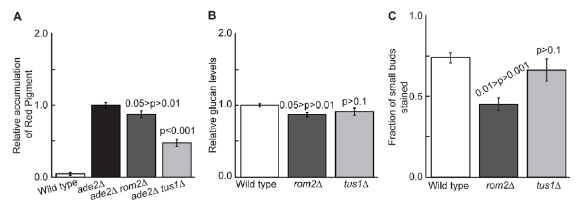
Rom2 appears to have more of a role in the bud growth with its localization coinciding with the bud tip, eventually, localizing to the contractile ring. However, Tus1 is localized at the contractile ring only. We show that Rom2 localization and partial function are dependent upon an additional protein that we, previously shown, to act upstream of Pkc1; the protein Ack1. Similarly, we discovered that Tus1 has a similar protein that helps navigate its functions, a protein we named Rgl1.

This work was highlighted by the Journal of Cell Science as one of their “In this issue” section.
Back to Drosophila.
My research with Prof. Julian Dow has involved creating resources for the Drosophila research community.
FlyAtlas2.
The first was an updated version of Fly Atlas, where we generated a sex-specific, tissue-specific transcriptome that included microRNAs. The website is here: https://motif.mvls.gla.ac.uk/FlyAtlas2/?page=gene#

The Drosophila transcriptomic data can be searched by genes, tissue, and category. With the category search, one can use GO terms as well as keywords. One can also search for genes that have a similar expression pattern to a particular gene of interest under the profile page.
Our FlyAtlas 2 data is incorporated into the Drosophila model organism go-to website, FlyBase.
Our next project was doing a similar thing as FlyAtlas2 but determining the metabolomes for each tissues in a sex-specific manner. Our data can be examined here: http://flymet.org/met_explore/
FlyMet.
Our next project was doing a similar thing as FlyAtlas2 but determining the metabolomes for each tissues in a sex-specific manner. Our data can be examined here: http://flymet.org/met_explore/

With FlyMet, one can search for different metabolites, peaks, pathways, and genes.
The most difficult aspect of using metabolomes in biology I found is that it is not looking at metabolite changes but changes in peaks. Also, just because metabolites are not present does not necessarily mean they are not relevant for that particular tissue.
We found it is easier to conceptualize what is happening with metabolome data by combining it with the analysis of transcriptome data.
Single Nuclei Transcriptomics.
Our current project involves a comparative anatomy/physiology of representative insects across the insect world. Our focus is their renal system and their ability to maintain water homeostasis. We are using single-cell technology to show how the various insects are similar and how they differ.
Committee work.
University of Glasgow-wide Postdoctoral Researcher Forum: 2015-2023 (when it ended)
College of MVLS ERC committee (NERD): 2023-present
School of MB ERC committee: 2019-present
School of MB EDI committee: 2022-present


