Professor Ilan Davis
- Professor of Spatial Molecular Biology (Molecular Biosciences)
- Affiliate Professor (School of Infection & Immunity)
telephone:
07941488029
email:
Ilan.Davis@glasgow.ac.uk
pronouns:
He/him/his
Personal assistant: Mr Jack Richardson
telephone: 01413303315
email: Jack.Richardson@glasgow.ac.uk
Biography
Ilan received his DPhil at ICRF Developmental Biology unit, Oxford University, after completing a Natural Sciences degree at Cambridge University. He did his postdoc at UCSF, San Francisco and then established his own lab at Edinburgh University in 1996 as a Wellcome Fellow, where he became a professor in 2003. He moved to Oxford university in 2007 to take up a professorship in Cell Biology, as a Wellcome Senior Fellow.
Ilan has been at the forefront of demonstrating that pattern formation, neural stem cell development and synaptic plasticity, involve crucial regulation by post-transcriptional regulation. His lab discovered the first example of Dynein transporting mRNA along microtubules during pattern formation in Drosophila embryo and oocyte and demonstrated that mRNA is statically anchored by the motor. They also characterised how phase transition processing bodies triage the fate of mRNA for immediate translation in oocytes or storage for later translation in the embryo.
More recently, they discovered a key step in regulating the rate of division and size of neural stem cells, involving the regulation of mRNA stability by an mRNA binding protein. They also discovered key steps in synaptic plasticity, the molecular basis of memory and learning, involving localised translation of actin binding regulators by another mRNA binding protein.
These studies have involved interdisciplinary work and the development of novel advanced microscopy, image analysis and computational biology methods. Most recently, the lab has also applied their unique expertise to highly sensitive detection of mRNA viruses, allowing the detection of viral replication earlier than previously possible.
Ilan was elected as an EMBO member in 2010.
Ilan's Davis's Lab: Dr Jeff Lee, Dr Sofia Polcownuk, Dr Danail Stoychev, Mrs Julia Dunlop (part-time)
Research interests
DROSOPHILA NEUROSCIENCE

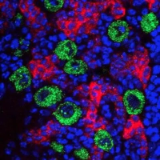
Synaptic Plasticity Neural Stem Cell Development

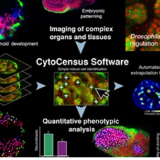
smFISH Screen Data Management and Analysis
VIRUSES AND RNA
Sars Cov2 Dynamics of Viral Infection RNA Biology & Method Development
MICROSCOPE TECHNOLOGY DEVELOPMENT


Microscopy Bespoke Microscope Building

Python Microscope Cockpit Image Analysis Software
Publications
Prior publications
Edited Book
(2017) RNA Detection: Methods and Protocols SPRINGER Ilan Davis.
Article
(2017) Brain Development: Machine Learning Analysis Of Individual Stem Cells In Live 3D Tissue Ilan Davis. (doi: https://doi.org/10.1101/137406 )
(2017) Imp/Syp Temporal Gradients Govern Decommissioning Of Drosophila Neural Stem Cells Ilan Davis. (doi: https://doi.org/10.1101/136655 )
(2017) Regulating prospero mRNA Stability Determines When Neural Stem Cells Stop Dividing Ilan Davis. (doi: https://doi.org/10.1101/135848 )
(2017) Single Molecule Fluorescence In Situ Hybridisation For Quantitating Post-Transcriptional Regulation In Drosophila Brains Ilan Davis. (doi: https://doi.org/10.1101/128785 )
(2016) Drosophila sensory cilia lacking MKS-proteins exhibit striking defects in development but only subtle defects in adults. Journal of Cell Science Ilan Davis.
Misra, M. et al. (2016) A genome-wide screen for dendritically localized RNAs identifies genes required for dendrite morphogenesis G3: Genes, Genomes, Genetics Scopus - Elsevier. (doi: 10.1534/g3.116.030353)
Pratt, M.B., Titlow, J.S., Davis, I., Barker, A.R., Dawe, H.R., Raff, J.W., Roque, H. (2016) Drosophila sensory cilia lacking MKS proteins exhibit striking defects in development but only subtle defects in adults Journal of Cell Science Scopus - Elsevier. (doi: 10.1242/jcs.194621)
Davidson, A., Parton, R.M., Rabouille, C., Weil, T.T., Davis, I. (2016) Localized Translation of gurken/TGF-α mRNA during Axis Specification Is Controlled by Access to Orb/CPEB on Processing Bodies Cell Reports Scopus - Elsevier. (doi: 10.1016/j.celrep.2016.02.038)
York-Andersen, A.H., Parton, R.M., Bi, C.J., Bromley, C.L., Davis, I., Weil, T.T. (2015) A single and rapid calcium wave at egg activation in Drosophila Biology Open Scopus - Elsevier. (doi: 10.1242/bio.201411296)
Johnson, E., Seiradake, E., Jones, E.Y., Davis, I., Grünewald, K., Kaufmann, R. (2015) Correlative in-resin super-resolution and electron microscopy using standard fluorescent proteins Scientific Reports Scopus - Elsevier. (doi: 10.1038/srep09583)
Ball, G., Demmerle, J., Kaufmann, R., Davis, I., Dobbie, I.M., Schermelleh, L. (2015) SIMcheck: A toolbox for successful super-resolution structured illumination microscopy Scientific Reports Scopus - Elsevier. (doi: 10.1038/srep15915)
McDermott, S.M., Yang, L., Halstead, J.M., Hamilton, R.S., Meignin, C., Davis, I. (2014) Drosophila Syncrip modulates the expression of mRNAs encoding key synaptic proteins required for morphology at the neuromuscular junction RNA Scopus - Elsevier. (doi: 10.1261/rna.045849.114)
Parton, R.M., Davidson, A., Davis, I., Weil, T.T. (2014) Subcellular mRNA localisation at a glance Journal of Cell Science Scopus - Elsevier. (doi: 10.1242/jcs.114272)
Kaufmann, R., Schellenberger, P., Seiradake, E., Dobbie, I.M., Jones, E.Y., Davis, I., Hagen, C., Grünewald, K. (2014) Super-resolution microscopy using standard fluorescent proteins in intact cells under cryo-conditions Nano Letters Scopus - Elsevier. (doi: 10.1021/nl501870p)
Halstead, J.M., Lin, Y.Q., Durraine, L., Hamilton, R.S., Ball, G., Neely, G.G., Bellen, H.J., Davis, I. (2014) Syncrip/hnRNP Q influences synaptic transmission and regulates BMP signaling at the Drosophila neuromuscular synapse Biology Open Scopus - Elsevier. (doi: 10.1242/bio.20149027)
McDermott, S.M., Davis, I. (2013) Drosophila Hephaestus/Polypyrimidine Tract Binding Protein Is Required for Dorso-Ventral Patterning and Regulation of Signalling between the Germline and Soma PLoS ONE Scopus - Elsevier. (doi: 10.1371/journal.pone.0069978)
McDermott, S.M., Meignin, C., Rappsilber, J., Davis, I. (2012) Drosophila Syncrip binds the gurken mRNA localisation signal and regulates localised transcripts during axis specification Biology Open Scopus - Elsevier. (doi: 10.1242/bio.2012885)
Weil, T.T. et al. (2012) Drosophila patterning is established by differential association of mRNAs with P bodies Nature Cell Biology Scopus - Elsevier. (doi: 10.1038/ncb2627)
Weil, T.T., Parton, R.M., Davis, I. (2012) Preparing individual Drosophila egg chambers for live imaging Journal of Visualized Experiments Scopus - Elsevier. (doi: 10.3791/3679)
Weil, T.T., Parton, R.M., Davis, I. (2012) Preparing individual Drosophila egg chambers for live imaging. Journal of visualized experiments : JoVE Scopus - Elsevier.
Hartswood, E., Brodie, J., Vendra, G., Davis, I., Finnegan, D.J. (2012) RNA:RNA interaction can enhance RNA localization in Drosophila oocytes RNA Scopus - Elsevier. (doi: 10.1261/rna.026674.111)
Brown, A.C.N., Dobbie, I.M., Alakoskela, J.-M., Davis, I., Davis, D.M. (2012) Super-resolution imaging of remodeled synaptic actin reveals different synergies between NK cell receptors and integrins Blood Scopus - Elsevier. (doi: 10.1182/blood-2012-05-429977)
Zhang, F. et al. (2012) UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery Cell Scopus - Elsevier. (doi: 10.1016/j.cell.2012.09.040)
Parton, R.M., Hamilton, R.S., Ball, G., Yang, L., Cullen, C.F., Lu, W., Ohkura, H., Davis, I. (2011) A PAR-1-dependent orientation gradient of dynamic microtubules directs posterior cargo transport in the Drosophila oocyte Journal of Cell Biology Scopus - Elsevier. (doi: 10.1083/jcb.201103160)
Hamilton, R.S., Davis, I. (2011) Identifying and searching for conserved RNA localisation signals. Methods in molecular biology (Clifton, N.J.) Scopus - Elsevier. (doi: 10.1007/978-1-61779-005-8_27)
Dobbie, I.M., King, E., Parton, R.M., Carlton, P.M., Sedat, J.W., Swedlow, J.R., Davis, I. (2011) OMX: A new platform for multimodal, multichannel wide-field imaging Cold Spring Harbor Protocols Scopus - Elsevier. (doi: 10.1101/pdb.top121)
Brown, A.C.N. et al. (2011) Remodelling of cortical actin where lytic granules dock at Natural Killer cell immune synapses revealed by super-resolution microscopy PLoS Biology Scopus - Elsevier. (doi: 10.1371/journal.pbio.1001152)
Yang, L., Parton, R., Ball, G., Qiu, Z., Greenaway, A.H., Davis, I., Lu, W. (2010) An adaptive non-local means filter for denoising live-cell images and improving particle detection Journal of Structural Biology Scopus - Elsevier. (doi: 10.1016/j.jsb.2010.06.019)
Oliveira, R.A., Hamilton, R.S., Pauli, A., Davis, I., Nasmyth, K. (2010) Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei Nature Cell Biology Scopus - Elsevier. (doi: 10.1038/ncb2018)
Parton, R.M., Vallés, A.M., Dobbie, I.M., Davis, I. (2010) Collection and mounting of drosophila embryos for imaging Cold Spring Harbor Protocols Scopus - Elsevier. (doi: 10.1101/pdb.prot5403)
Weil, T.T., Xanthakis, D., Parton, R., Dobbie, I., Rabouille, C., Gavis, E.R., Davis, I. (2010) Distinguishing direct from indirect roles for bicoid mRNA localization factors Development Scopus - Elsevier. (doi: 10.1242/dev.044867)
Parton, R.M., Vallés, A.M., Dobbie, I.M., Davis, I. (2010) Drosophila larval fillet preparation and imaging of neurons Cold Spring Harbor Protocols Scopus - Elsevier. (doi: 10.1101/pdb.prot5405)
Parton, R.M., Vallés, A.M., Dobbie, I.M., Davis, I. (2010) Drosophila macrophage preparation and screening Cold Spring Harbor Protocols Scopus - Elsevier. (doi: 10.1101/pdb.prot5404)
Parton, R.M., Davis, I. (2010) How the sea squirt nucleus tells mesoderm not to be endoderm Developmental Cell Scopus - Elsevier. (doi: 10.1016/j.devcel.2010.10.002)
Parton, R.M., Vallés, A.M., Dobbie, I.M., Davis, I. (2010) Isolation of Drosophila egg chambers for imaging Cold Spring Harbor Protocols Scopus - Elsevier. (doi: 10.1101/pdb.prot5402)
Weil, T.T., Parton, R.M., Davis, I. (2010) Making the message clear: Visualizing mRNA localization Trends in Cell Biology Scopus - Elsevier. (doi: 10.1016/j.tcb.2010.03.006)
Hamilton, R.S., Parton, R.M., Oliveira, R.A., Vendra, G., Ball, G., Nasmyth, K., Davis, I. (2010) ParticleStats: Open source software for the analysis of particle motility and cytoskeletal polarity Nucleic Acids Research Scopus - Elsevier. (doi: 10.1093/nar/gkq542)
Meignin, C., Davis, I. (2010) Transmitting the message: intracellular mRNA localization Current Opinion in Cell Biology Scopus - Elsevier. (doi: 10.1016/j.ceb.2009.11.011)
Hamilton, R.S., Hartswood, E., Vendra, G., Jones, C., Van De Bor, V., Finnegan, D., Davis, I. (2009) A bioinformatics search pipeline, RNA2DSearch, identifies RNA localization elements in Drosophila retrotransposons RNA Scopus - Elsevier. (doi: 10.1261/rna.1264109)
Pickard, B.S. et al. (2008) A common variant in the 3′UTR of the GRIK4 glutamate receptor gene affects transcript abundance and protects against bipolar disorder Proceedings of the National Academy of Sciences of the United States of America Scopus - Elsevier. (doi: 10.1073/pnas.0800643105)
Weil, T.T., Parton, R., Davis, I., Gavis, E.R. (2008) Changes in bicoid mRNA Anchoring Highlight Conserved Mechanisms during the Oocyte-to-Embryo Transition Current Biology Scopus - Elsevier. (doi: 10.1016/j.cub.2008.06.046)
Zimyanin, V.L., Belaya, K., Pecreaux, J., Gilchrist, M.J., Clark, A., Davis, I., St Johnston, D. (2008) In Vivo Imaging of oskar mRNA Transport Reveals the Mechanism of Posterior Localization Cell Scopus - Elsevier. (doi: 10.1016/j.cell.2008.06.053)
Meignin, C., Davis, I. (2008) UAP56 RNA helicase is required for axis specification and cytoplasmic mRNA localization in Drosophila Developmental Biology Scopus - Elsevier. (doi: 10.1016/j.ydbio.2007.12.004)
Clark, A., Meignin, C., Davis, I. (2007) A Dynein-dependent shortcut rapidly delivers axis determination transcripts into the Drosophila oocyte Development Scopus - Elsevier. (doi: 10.1242/dev.02832)
Delanoue, R., Herpers, B., Soetaert, J., Davis, I., Rabouille, C. (2007) Drosophila Squid/hnRNP Helps Dynein Switch from a gurken mRNA Transport Motor to an Ultrastructural Static Anchor in Sponge Bodies Developmental Cell Scopus - Elsevier. (doi: 10.1016/j.devcel.2007.08.022)
Vendra, G., Hamilton, R.S., Davis, I. (2007) Dynactin suppresses the retrograde movement of apically localized mRNA in Drosophila blastoderm embryos RNA Scopus - Elsevier. (doi: 10.1261/rna.509007)
Tekotte, H., Tollervey, D., Davis, I. (2007) Imaging the migrating border cell cluster in living Drosophila egg chambers Developmental Dynamics Scopus - Elsevier. (doi: 10.1002/dvdy.21305)
Hamilton, R.S., Davis, I. (2007) RNA localization signals: Deciphering the message with bioinformatics Seminars in Cell and Developmental Biology Scopus - Elsevier. (doi: 10.1016/j.semcdb.2007.02.001)
Meignin, C., Alvarez-Garcia, I., Davis, I., Palacios, I.M. (2007) The Salvador-Warts-Hippo Pathway Is Required for Epithelial Proliferation and Axis Specification in Drosophila Current Biology Scopus - Elsevier. (doi: 10.1016/j.cub.2007.09.062)
Tekotte, H., Davis, I. (2006) Bruno: A double turn-off for oskar Developmental Cell Scopus - Elsevier. (doi: 10.1016/j.devcel.2006.02.001)
Delanoue, R., Davis, I. (2005) Dynein anchors its mRNA cargo after apical transport in the Drosophila blastoderm embryo Cell Scopus - Elsevier. (doi: 10.1016/j.cell.2005.04.033)
Van De Bor, V., Hartswood, E., Jones, C., Finnegan, D., Davis, I. (2005) gurken and the I factor retrotransposon RNAs share common localization signals and machinery Developmental Cell Scopus - Elsevier. (doi: 10.1016/j.devcel.2005.04.012)
Davis, I. (2004) A helicase that gets Oskar's message across Nature Cell Biology Scopus - Elsevier. (doi: 10.1038/ncb0404-285)
Van De Bor, V., Davis, I. (2004) mRNA localisation gets more complex Current Opinion in Cell Biology Scopus - Elsevier. (doi: 10.1016/j.ceb.2004.03.008)
MacDougall, N., Clark, A., MacDougall, E., Davis, I. (2003) Drosophila gurken (TGFα) mRNA localizes as particles that move within the oocyte in two dynein-dependent steps Developmental Cell Scopus - Elsevier. (doi: 10.1016/S1534-5807(03)00058-3)
Tekotte, H., Berdnik, D., Török, T., Buszczak, M., Jones, L.M., Cooley, L., Knoblich, J.A., Davis, I. (2002) Dcas is required for importin-α3 nuclear export and mechano-sensory organ cell fate specification in Drosophila Developmental Biology Scopus - Elsevier. (doi: 10.1006/dbio.2002.0612)
Tekotte, H., Davis, I. (2002) Intracellular mRNA localization: Motors move messages Trends in Genetics Scopus - Elsevier. (doi: 10.1016/S0168-9525(02)02819-6)
Davis, I. (1997) Nuclear polarity and nuclear trafficking in Drosophila Seminars in Cell and Developmental Biology Scopus - Elsevier.
Francis-Lang, H., Davis, I., Ish-Horowicz, D. (1996) Asymmetric localization of Drosophila pair-rule transcripts from displaced nuclei: Evidence for directional nuclear export EMBO Journal Scopus - Elsevier.
Davis, I., Girdham, C.H., O'Farrell, P.H. (1995) A Nuclear GFP That Marks Nuclei in Living Drosophila Embryos; Maternal Supply Overcomes a Delay in the Appearance of Zygotic Fluorescence Developmental Biology Scopus - Elsevier. (doi: 10.1006/dbio.1995.1251)
Davis, I., Francis-Lang, H., Ish-Horowicz, D. (1993) Mechanisms of intracellular transcript localization and export in early Drosophila embryos Cold Spring Harbor Symposia on Quantitative Biology Scopus - Elsevier.
Davis, I., Ish-Horowicz, D. (1991) Apical localization of pair-rule transcripts requires 3′ sequences and limits protein diffusion in the Drosophila blastoderm embryo Cell Scopus - Elsevier.
Book
Waithe, D., Hailstone, M., Lalwani, M.K., Parton, R., Yang, L., Patient, R., Eggeling, C., Davis, I. (2016) 3-D density kernel estimation for counting in microscopy image volumes using 3-D image filters and random decision trees Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) Scopus - Elsevier. (doi: 10.1007/978-3-319-46604-0_18)
Ball, G., Parton, R.M., Hamilton, R.S., Davis, I. (2012) A cell biologist's guide to high resolution imaging Methods in Enzymology Scopus - Elsevier. (doi: 10.1016/B978-0-12-391857-4.00002-1)
Parton, R.M., Davis, I. (2006) Lifting the Fog: Image Restoration by Deconvolution Cell Biology, Four-Volume Set Scopus - Elsevier. (doi: 10.1016/B978-012164730-8/50147-7)
Conference Proceedings
Yang, L., Parton, R., Ball, G., Qiu, Z., Greenaway, A.H., Davis, I., Lu, W. (2010) A 2D+t feature-preserving non-local means filter for image denoising and improved detection of small and weak particles British Machine Vision Conference, BMVC 2010 - Proceedings Scopus - Elsevier. (doi: 10.5244/C.24.14)
Grants
Grants and Awards listed are those received whilst working with the University of Glasgow.
- Democratising spatial transcriptomics
Biotechnology and Biological Sciences Research Council
2025 - 2026
- Mechanistic analysis of TDP-43-mediated RNA localization in neurons and its misregulation in ALS
National Institutes of Health
2023 - 2024
- Mechanistic analysis of TDP-43-mediated RNA localization in neurons and its misregulation in ALS
National Institutes of Health
2023 - 2025
Supervision
- Lavery, Jessica
N/A - Qin, Zhiling
The host and viral factors influencing SARS-CoV2 infection and replication dynamics in culture models and human patient samples



