Lab/infrastructure
Dr Konstantinos Gerasimidis' Lab
The BINGO group at the University of Glasgow hosts a broad range of expertise and resources aiming to study the role of diet and its interaction with the gut microbiome in health and disease; focusing predominantly in human-based trials. Dr Gerasimidis group is located in a containment Class 2 laboratory facility based in the New Lister Building, at the Glasgow Royal Infirmary. Dr Gerasimidis’ group is equipped with mainstream molecular microbiology, food analysis, and analytical chromatographic techniques which enable profiling of the gut microbiome and measurement of a broad spectrum of analytes in biological samples. Dietary interventions and metabolic studies in humans take place in-situ, in dedicated and purpose-built laboratory kitchens, energy-balance laboratories and facilities to collect and analyse biological samples. Adjacent to this facility are our dedicated Clinical Research Facilities operating under the National Health Service in Scotland (http://www.nhsresearchscotland.org.uk/research-in-scotland/facilities/clinical-research-facilities).


Professor Simon Milling Lab
The Centre of Immunobiology, within the institute of Infection, Immunity & Inflammation, occupies three floors (each with 250m2 of laboratory space) of the purpose-built Sir Graeme Davies Building, including the flow core facility (www.gla.ac.uk/researchinstitutes/iii/facilities/fcf), for which Prof Milling is the Academic Lead. This is equipped with state-of-the-art equipment including two FACS Aria III cell sorters, a BD LSRFortessa™, and three other multi-laser flow cytometers, managed by a dedicated highly experienced full-time member of staff.
The project team also receives support from the group’s Clinical Research Dietitian and Clinical Research Nurse.
Dr Umer Zeeshan Ijaz’s Lab
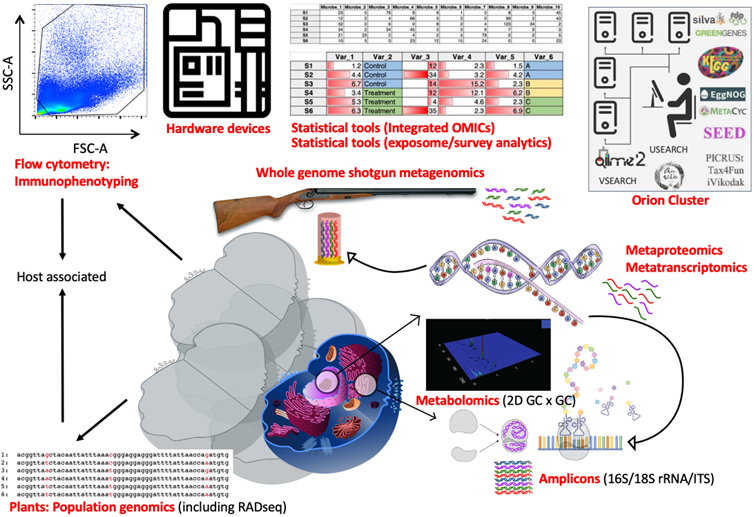
- Research Environment: Dr Ijaz’s Environmental’Omics lab is part of Water & Environment (W&E) Group. Since 2022, W&E group by virtue of its repute is assimilated in Global Sustainable Development Theme and is hosted in a £116M Mazumdar-Shaw Advanced Research Centre which has also been awarded the 2022 Pupil/Student Experience of the Year by Learning Places Scotland. In addition to the state-of-the-art facilities provided by the research centre, W&E group provides Environmental Biotechnology Laboratories to carry out a wet-lab activities for microbial community surveys using DNA, flowcytometry and gas chromatography.
- Hardware Infrastructure: Dr Ijaz’s lab supports bioinformatics HPC facility, Orion Cluster (~500 cores; ~800TB disk space), that has over 500 bioinformatics tools and bespoke workflows, specifically tailored for "big data" projects. Additionally, Dr Ijaz has released the bespoke software and workflows, some of these also run on Orion cluster: CONCOCT (2014; one of the first international attempts at segregating microbial genomes; DOI: 10.1038/nmeth.3103), RVLAB (2016; an online statistical processing environment for multivariate analysis of microbial communities; 10.3897/BDJ.4.e8357); NMGS (2017; a software for fitting unified theory of neutral models to microbial communities; DOI: 10.1109/JPROC.2015.2428213); SEQENV & SEQENV-EXT (2016-2017; software for text mining environmental ontology terms, habitat identification from online text sources associated with genomic sequences; DOI: 10.7717/peerj.2690; DOI: 10.7717/peerj.3827); microbiomeSEQ (2017 - present; an R package for microbial community analysis in an environmental context; DOI: 10.13140/RG.2.2.17108.71047); NanoAmpli-Seq (2019; a workflow for amplicon sequencing from mixed microbial communities (DOI: 10.1093/gigascience/giy140); PyTag (2018; a software facilitating systematic reviews using prevalent ontologies (DOI: 10.7717/peerj.5047); Meta-Analysis Workflow (2020; strategy to collate and analyse publicly available microbiome datasets; DOI: 10.1101/2020.12.23.424166) and CViewer (2024; extension of CONCOCT; a software for multivariate statistical analysis of shotgun metagenomics datasets with other OMICS modalities; DOI: 10.1101/2023.06.07.544017). These software are modular in design, and collectively form a suite of software/workflows that interpret and process the massive amount of data that emerge from powerful high throughput molecular methods and omics technologies.
- Software Infrastructure and Training Activities: Dr Ijaz’s lab has developed bespoke training modules that include video lectures on processing data from omics technologies. These are widely in use (>70 visitors since 2014), and in the past, several visitors were successful in acquiring fundings through mobility schemes for placement in his lab, e.g., European Union's Earth System Science and Environmental Management ES1103 COST Action, Pakistan Higher Education Commission’s International Research Support Inititive Programme, Marie-Curie Programme, Austrian Erwin Schrodinger Fellowship, Encyclopedia of Life 2013 Rubenstein Research Fellowship, Research Council of Norway KLIMAFORSK Mobility Grant, Erasmus+ International Credit Mobility, and Charles Wallace Visiting Fellowships. The training modules include analysis of microbial community datasets using both short-read amplicons (16S/18S rRNA) and whole genome shotgun sequencing. Furthermore, multivariate statistical analyses (ecological statistics) in R for data generated from omics technologies is covered as well as given experience on writing bespoke scripts in bash/awk and python to process the data in an HPC environment.