New Paper led by Alisha Aman
Published: 11 January 2023
Investigating the potential impact of PCSK9-inhibitors on mood disorders using eQTL-based Mendelian randomization
Investigating the potential impact of PCSK9-inhibitors on mood disorders using eQTL-based Mendelian randomization
Alisha Aman ,Eric A. W. Slob,Joey Ward,Breda Cullen,Nicholas Graham,Donald M. Lyall,Naveed Sattar,Rona J. Strawbridge
Introduction
Cardiovascular disease (CVD) is currently the leading cause of mortality in the world with 32% of global deaths attributed to it in 2019 [1]. Low-density lipoprotein cholesterol (LDL-C) is one of the key causal risk factors for CVD and a range of potent treatments such as statins reduce LDL-C level in the blood and lower CVD outcomes. A more recent LDL-C treatment are the PCSK9-inhibitors (PCSK9i), and also lower CVD outcomes. The PCSK9 gene is located on chromosome 1 and encodes the proprotein convertase subtilisin/kexin type 9 (PCSK9) protein whose function is to target LDL-C receptors (LDLR) for degradation. Certain mutations in the PCSK9 gene lead to excess removal of LDL-C receptors which in turn lowers the uptake of LDL-C into the cell and ultimately lead to higher levels of LDL-C in the blood. PCSK9i are usually monoclonal antibodies that block the excess PCSK9, preventing the degradation of LDL-C receptors, thereby lowering circulating LDL-C levels [2]. PCSK9i are prescribed for the treatment of familial hypercholesterolemia or in patients with hypercholesterolemia and atherosclerotic CVD. They are also used in conjunction with statins or other lipid-lowering medications for additional reduction of LDL-C in blood [3]. Recently, price reduction of PCSK9i has led to an increase in prescription amongst eligible patients [4]. However, there is little information on potential adverse drug reaction (ADR) or off-target effects of PCSK9i.
Materials and methods
Exposure data
eQTLs are genetic variants which have genotype-specific effects on levels of the gene’s expression [18]. The principle behind our analyses plan is that the alleles for an eQTL which reduce the expression level of the gene, leading to lesser gene product, can be used as a proxy for the effect of a drug that reduces the level of the same gene product. Hence, we can use gene-expression level as the exposure to find the causal association with the outcomes of interest. Only cis-eQTLs, which are eQTLs within 1Mb window of the gene and on the same chromosome [18], were used in this study as the instrumental variables (IVs) for the MR analyses.
Outcome data
GWAS summary statistics with the largest sample sizes in European ancestry individuals were extracted using the IEUGWAS R package [23]. Outcomes considered were mood instability (id: ukb-b-14180, n = 451,619), neuroticism score (id: ukb-b-4630, n = 374,323), MDD (id: ebi-a-GCST005903, n = 217,584) [23, 24]. For positive controls, we used LDL-C (id: ieu-b-110, n = 403,943) [23, 24], and circulating PCSK9 levels (n = 12,721) [25]. Information on the GWAS studies used is detailed in S1 Table. If no GWAS summary statistic was available for an eQTL, data from its proxy at r2 threshold of 0.8 was used wherever possible. The cis-eQTL and GWAS summary statistics were combined, harmonized to the PCSK9 gene-expression reducing allele.
Results
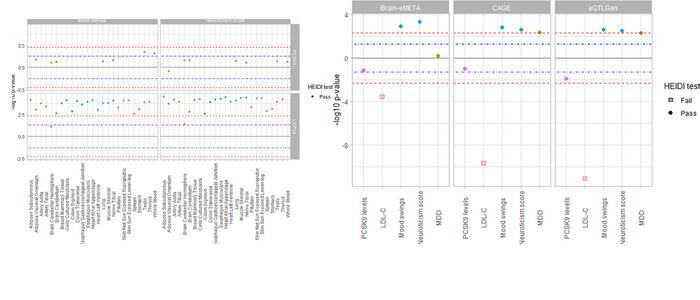
PCSK9 gene-expression is unlikely to be associated with mood disorders
SMR analysis was used to assess the association of PCSK9 gene-expression with PCSK9 and LDL-C levels (as positive controls) as well as MDD, mood instability, and neuroticism score. The expected associations with PCSK9-expression and LDL-C levels were observed, and there was a suggestive association between PCSK9-expression and mood instability in testis (pSMR_multi = 0.018, pHEIDI = 0.07), and whole blood (pSMR_multi = 0.02, pHEIDI = 0.18), but this did not reach significance after multiple testing correction. No significant associations were observed between PCSK9 gene-expression, and MDD, or neuroticism score (Fig 1 and S1 Fig, S3 Table). Our analyses were also sufficiently powered, mainly due to the use of outcome GWAS summary statistics from UK BioBank cohort, which has a very large sample size.
First published: 11 January 2023
Related Links
Article Information

