Brierley Group
Research Overview
The VIBE (Viral Informatics, Biostatistics, and Evolution) lab uses newly emerging data and computational methods to investigate what traits drive RNA viruses to ‘host shift’ (i.e., transmit to, infect, and cause disease within a new host species) and how such traits evolve, with a view to anticipating future pandemic risks.
Current Research
We use a broad range of methodologies (e.g., biostatistics, artificial intelligence, natural language processing) and model a broad diversity of animal RNA viruses, though our research is driven by consistent key questions around dynamics of viruses in their…
…ecological contexts
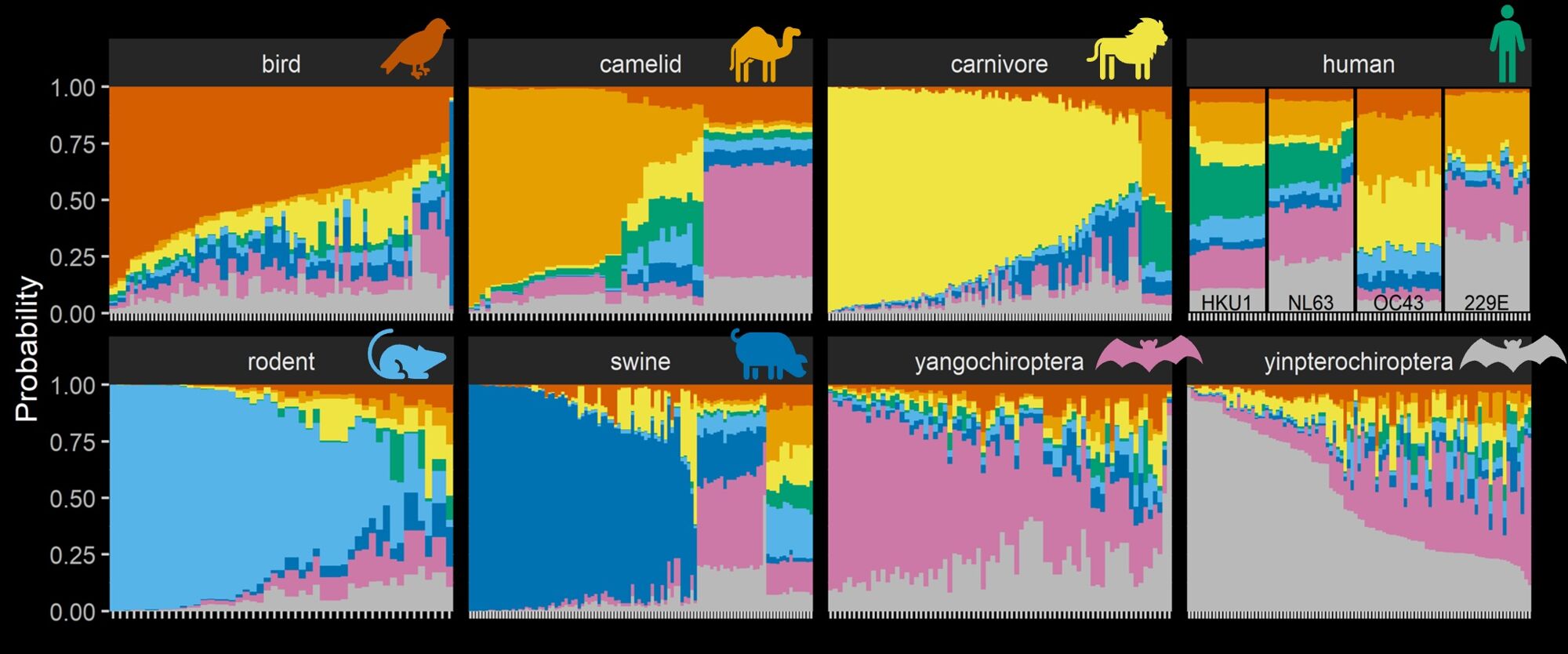
If we are to anticipate future human disease outbreaks, we need to better understand dynamics in wild hosts and the environments in which we interact with them. We are interested in modelling diversity of animal hosts and changes to landscapes influence cross-species transmission and emergence in humans, including global distributions of bat-borne viruses and the ecological shifts associated with the 2021 outbreak of highly pathogenic avian influenza.
…evolutionary dynamics
Evolutionary adaptation is the driving force underlying host range of a virus. A core theme of our research is understanding how ability to infect specific hosts and tissues can be characterised directly from genome sequences by training artificial intelligence approaches on genomic and proteomic traits (‘‘genotype-to-phenotype’’) - and how these traits evolve over time. We are also interested in whether evolutionary models can highlight viruses likely to be transmissible or virulent in human populations. Being at the intersection of evolution and computer science, our work often necessitates development of new computational methods for viral sequence data.
…public health impact
Although human populations are continuously exposed to novel viruses, few emerge successfully to cause significant outbreaks. Through collaborations, we are exploring how genomic and machine learning methodologies can improve predicted impact of emerging viruses on public health. This includes modelling chance of discovering new viruses, dynamics of transmissibility within populations, and outbreak characteristics.
…systematic data gaps
Our collective scientific knowledge of how viruses interact with their hosts is vast, although systematic data gaps exist. Our group is dedicated to understanding these gaps through statistical analysis and resolving them with modern computational tools. A primary focus of our current research is text mining to unlock the huge volume of information on host-parasite relationships in scientific literature. We are exploring natural language processing and large language model methods to generate usable data resources describing viral interactions with hosts, tissues, proteins, and more.
Group Members
|
Liam Brierley |
Jordon Bone |
Collaborators
- Dr Maxwell Farrell (SBOVHM, University of Glasgow)
- Dr Sarah Hayes (University of Oxford)
- Christl Donnelly (University of Oxford)
- Dr Jasmina Panovska-Griffiths (University of Oxford)
- Matthew Baylis (University of Liverpool)
- Iain Buchan (University of Liverpool)
- Dr Jan Gogarten (Helmholtz Institute for One Health)
- Dr Sébastien Calvignac-Spencer (Helmholtz Institute for One Health)
- Dr Lu Lu (Roslin Institute, University of Edinburgh)