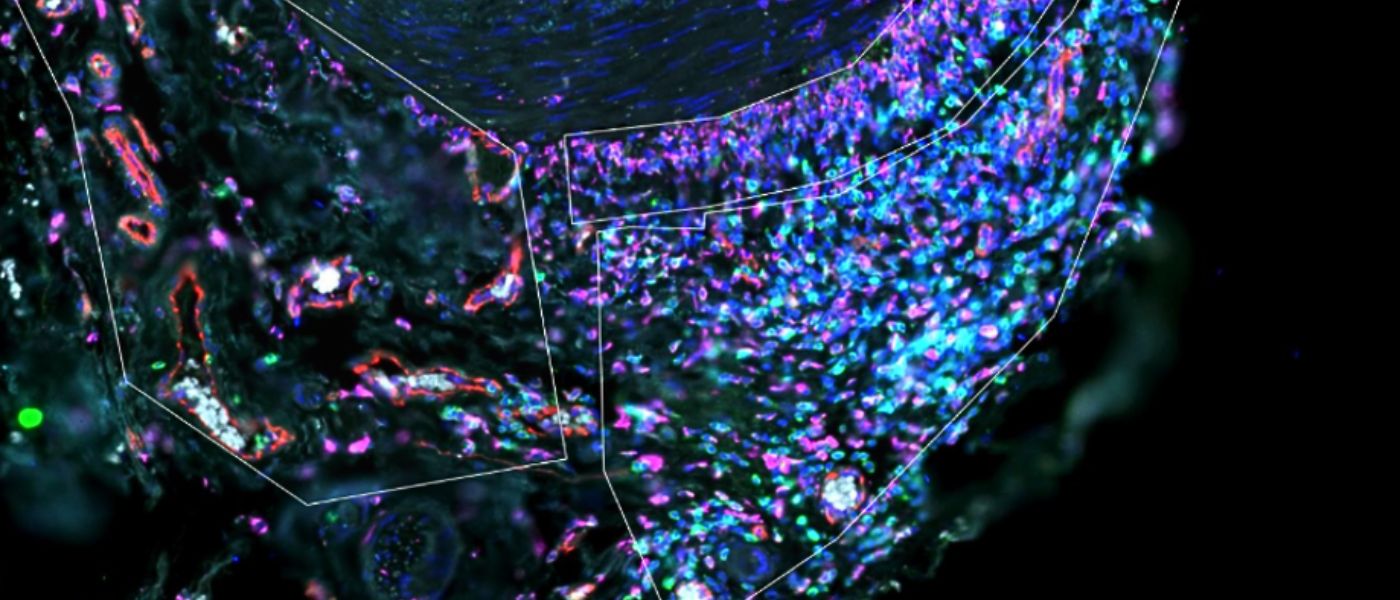
Spatial Transcriptomics - CYGNUS
The Living Laboratory's CYGNUS project is developing a powerful new pipeline for the detailed study of diagnostic and archival tissue samples. This will allow researchers to explore spatial (location) transcriptomic (RNA messages made by the cells) data which can tell us how cells interact and function in various environments.
By doing so, CYGNUS seeks to produce valuable, highly characterised datasets that can be used for wider exploration, fuelling further research and discovery.
In its initial phase, CYGNUS will provide spatial-omics support to five disease-specific exemplar projects. This will help define the pipeline and services needed to further expand and enhance the precision medicine research ecosystem within Glasgow.
Spatial Transcriptomics Explained
Spatial transcriptomics is a transformative technique that has revolutionised the way scientists observe gene expression within tissues. By visualising genes at a spatial level, this technology uncovers new pathways to understand the intricate relationships between genes, cells, and their surroundings.
The spatial organisation of cells within tissues is crucial for understanding biological processes and how cells interact with each other in their natural environment. This has significant implications for studying complex diseases and developmental biology.
The CYGNUS Project
In its initial phase, CYGNUS will work with both academic and industry collaborators to deliver five disease-specific exemplar projects.
1. Adenoid Cystic Carcinoma
Professor Ross Cagan
Exploring disease-associated pathways in Adenoid Cystic Carcinoma (ACC) to identify new drug targets and to validate a repositioning strategy that includes biomarker.
2. Inflammatory Bowel Disease
Dr David Bunton (Reprocell) and Dr Jonathan MacDonald (NHS)
Enhancing Reprocell’s commercial portfolio, understanding if spatial transcriptomics will allow for the exploration of small but vital signals to be uncovered in inflammatory bowel disease.
3. Non-small Cell Lung Cancer (NSCLC)
Professor Kevin Blyth
Creating a multi-modal spatial dataset using real world medical data from patients with non-small cell lung cancer, to be used for hypothesis testing and AI model generation by researchers and commercial companies.
4. Hepatobiliary Cancer
Professor Nigel Jamieson
Addressing the unmet clinical need in Pancreatico-biliary carcinomas, through the generation of a deeply phenotyped, single-cell spatial-omic dataset linked to H&E images that can be explored by third parties.
5. Giant Cell Arteritis
Professor Carl Goodyear and Dr Cecilia Ansalone
Identifying spatial-omic signatures for patient stratification and mechanistic pathways for drug targeting in Giant Cell Arteritis (GCA).
Outcomes
The CYGNUS project aims to demonstrate the significant value of the spatial transcriptomics pipeline within Glasgow. By building on existing partnerships and fostering new collaborations, CYGNUS will deliver exemplar projects that meet the specific needs of the NHS, biotech, pharma, and academia.
These exemplars will drive healthcare outcomes, such as improved patient stratification and the identification of novel targets and biomarkers. This will enhance the precision medicine landscape and create tangible benefits for both patients and researchers.